Introducing Your Microbes
What causes you to be how you are? The classic answer is a mixture of nature (your personal DNA) and nurture (your experiences within your environment). Until quite recently, scientific opinion basically agreed with this assessment. Now, however, the consensus is that the behaviour of our bodies depends on our microbes just as surely as it depends on our nature and nurture.
Welcome to the fascinating new world of the Human Microbiome.
Disclaimer: This is a tour of the scientific literature, not medical advice or an alternative to the opinion of your doctor. Please behave responsibly and use it for intellectual appreciation, not self-treatment.
The Human Microbiome
For centuries people have thought of microbes as things that cause disease. It’s only recently that we’ve realized they are also fundamental to our existence. We all have microbes on our skin, in our digestive systems and our reproductive systems. Almost every bodily system you can think of is a team effort: swarms of unimaginably many human and bacterial cells interacting selfishly and altruistically in the beautiful dance of life.
The ecosystem is your body, and the community of microbes that live in it is called the Human Microbiome. It’s an area of research that’s advancing at a tremendous pace, with study after study showing new links between our behavior and our microbes.
The American Society of Microbiology has produced a nice overview here.
Digestion
Microbes are a fundamental part of digestion. In fact, our digestive system contains the largest population of microbes in our body. Like our immune system it evolved to interact with organisms (in this case food) that evolve on shorter timespans than we do. By partnering with bacteria our stomachs can rapidly adapt to new foods and changing diets.
What we know:
-
We need microbes to fully digest most foods.
-
Normally functioning gut microbes serve as the first line of defense for the immune system. When properly functioning, they competitively exclude pathogens like Clostridium difficile by out-competing them for resources.
-
Your gastrointestinal tract has its own nervous system, the enteric nervous system (Scientific American version here), capable of releasing serotonin, dopamine and other psychologically active signaling chemicals. Microbially mediated release of these neurotransmitters is one of the causal mechanisms that serves as link between your microbiome and your happiness and stress levels.
-
Your stomach’s ecosystem is so complex that many scientists have come to think of it as an organ. An organ sadly misunderstood and mistreated throughout much of human history.
Here some relevant papers:
Prof. Gordon from WUSTL got a lot of people excited in 2006 when his team showed that obese mice have different gut microbes than skinny mice.
Full paper here, NPR summary here. A key paragraph from the paper:
Comparisons of the distal gut microbiota of genetically obese mice and their lean littermates, as well as those of obese and lean human volunteers have revealed that obesity is associated with changes in the relative abundance of the two dominant bacterial divisions, the Bacteroidetes and the Firmicutes. Here we demonstrate through metagenomic and biochemical analyses that these changes affect the metabolic potential of the mouse gut microbiota. Our results indicate that the obese microbiome has an increased capacity to harvest energy from the diet. Furthermore, this trait is transmissible: colonization of germ-free mice with an 'obese microbiota' results in a significantly greater increase in total body fat than colonization with a 'lean microbiota'.
Isn’t that cool? There are skinny microbiomes and fat microbiomes!
After Prof. Gordon’s results there was an explosion of interest. BGI-Shenzen surveyed European gut samples and found somewhere between 1,000 and 1,500 bacterial species. They made the first attempts at defining a minimal gut metagenome and its metabolism. It was the first large cohort of humans and their microbes, our first in-depth look into the world of the human microbiome. Full paper here.
Abstract: To understand the impact of gut microbes on human health and well-being it is crucial to assess their genetic potential. Here we describe the Illumina-based metagenomic sequencing, assembly and characterization of 3.3 million non-redundant microbial genes, derived from 576.7 gigabases of sequence, from faecal samples of 124 European individuals. The gene set, ~150 times larger than the human gene complement, contains an overwhelming majority of the prevalent (more frequent) microbial genes of the cohort and probably includes a large proportion of the prevalent human intestinal microbial genes. The genes are largely shared among individuals of the cohort. Over 99% of the genes are bacterial, indicating that the entire cohort harbours between 1,000 and 1,150 prevalent bacterial species and each individual at least 160 such species, which are also largely shared. We define and describe the minimal gut metagenome and the minimal gut bacterial genome in terms of functions present in all individuals and most bacteria, respectively.
For my postdoctoral research I joined the NIH’s Human Microbiome Project, which did an even larger survey, including different body parts. We not only discovered many species of gut microbes, but also mapped out the core metabolic capabilities of many distinct microbiomes all around the body. We found (among many other things!) that although two people may have drastically different collections of stomach flora, they tend to have shared metabolic capabilities. Here’s one of the main studies.
Abstract: Studies of the human microbiome have revealed that even healthy individuals differ remarkably in the microbes that occupy habitats such as the gut, skin and vagina. Much of this diversity remains unexplained, although diet, environment, host genetics and early microbial exposure have all been implicated. Accordingly, to characterize the ecology of human-associated microbial communities, the Human Microbiome Project has analysed the largest cohort and set of distinct, clinically relevant body habitats so far. We found the diversity and abundance of each habitat's signature microbes to vary widely even among healthy subjects, with strong niche specialization both within and among individuals. The project encountered an estimated 81-99% of the genera, enzyme families and community configurations occupied by the healthy Western microbiome. Metagenomic carriage of metabolic pathways was stable among individuals despite variation in community structure, and ethnic/racial background proved to be one of the strongest associations of both pathways and microbes with clinical metadata. These results thus delineate the range of structural and functional configurations normal in the microbial communities of a healthy population, enabling future characterization of the epidemiology, ecology and translational applications of the human microbiome.
Because we shared our data with the world, this study created an incredible number of follow-up works. Many people re-analyzed our data, finding new and interesting details in the microbiota.
Microbes are everywhere, doing essential things for us. If we have the wrong kind of microbes, or are missing microbes, we’re in trouble.
What determines your microbial make-up?
The short answer would be: many factors.
We have to look at both how we acquire new microbes, and what their lifecycles are. New microbes come from the sources you might expect: every lungful of air, each bite of food, every surface contact. Our every second on this planet brings with it new microbes. The microbes we encounter can have significant geographic variation (or not!, some microbes are very stable across multiple geographic variations).
A fundamental organizing fact here is that some microbes have generation times of about 20 minutes, instead of about 20 years like we do. However, bacteria are very diverse and some take days. Fast growers can colonize new places swiftly, but are sometimes outcompeted by slower growers after both have been around for a while. This can lead to an ecological process called “succession” when bacteria colonize humans. Another issue is that microbes can depend upon other microbes in their ecosystem, leading to complex dynamics.
As you move between cities, states, and countries, the mismatch between your current microbiome and what the environment says it should be can potentially lead to large scale mismatches that cause traveler’s diarrhea and other unfortunate outcomes.
Your behavior also has a tremendous effect on your microbiome. Showering, antibiotic treatments, and chlorinated water decimate their ranks. Your dietary choices, which lotion you use, and myriad other factors all drastically shift the nutrients available to your microbes.
Also, one observation that seems to be mostly true in microbiology is: “everything is everywhere, but the environment selects”. This suggests that given favorable enough conditions, we might get most microbes.
Yet, we don’t always get the same selection.
A look across different populations sheds some light on this; from the paper:
Abstract: Gut microbial communities represent one source of human genetic and metabolic diversity. To examine how gut microbiomes differ among human populations, here we characterize bacterial species in fecal samples from 531 individuals, plus the gene content of 110 of them. The cohort encompassed healthy children and adults from the Amazons of Venezuela, rural Malawi and US metropolitan areas and included mono- and dizygotic twins. Shared features of the functional maturation of the gut microbiome were identified during the first three years of life in all three populations, including age-associated changes in the genes involved in vitamin biosynthesis and metabolism. Pronounced differences in bacterial assemblages and functional gene repertoires were noted between US residents and those in the other two countries. These distinctive features are evident in early infancy as well as adulthood. Our findings underscore the need to consider the microbiome when evaluating human development, nutritional needs, physiological variations and the impact of westernization.
The way you use your body parts also has a large impact. One of the most dramatic examples comes from the work of Prof. Knight’s team at Univ. of Colorado, where they examined what microbes live on your hands. He showed that the similarities between the ecosystems of your two hands was only 18%. Why? It’s early, but the answer seems to be handedness. Differences in hand behavior had led to different hand ecosystems.
Abstract: Bacteria thrive on and within the human body. One of the largest human-associated microbial habitats is the skin surface, which harbors large numbers of bacteria that can have important effects on health. We examined the palmar surfaces of the dominant and nondominant hands of 51 healthy young adult volunteers to characterize bacterial diversity on hands and to assess its variability within and between individuals. We used a novel pyrosequencing-based method that allowed us to survey hand surface bacterial communities at an unprecedented level of detail. The diversity of skin-associated bacterial communities was surprisingly high; a typical hand surface harbored >150 unique species-level bacterial phylotypes, and we identified a total of 4,742 unique phylotypes across all of the hands examined. Although there was a core set of bacterial taxa commonly found on the palm surface, we observed pronounced intra- and interpersonal variation in bacterial community composition: hands from the same individual shared only 17% of their phylotypes, with different individuals sharing only 13%. Women had significantly higher diversity than men, and community composition was significantly affected by handedness, time since last hand washing, and an individual's sex. The variation within and between individuals in microbial ecology illustrated by this study emphasizes the challenges inherent in defining what constitutes a "healthy" bacterial community; addressing these challenges will be critical for the International Human Microbiome Project.
Moving to a smaller scale, differences between the regions of your body control microbial densities. Microbes suited to armpits do not reproduce as well on the lower back, and so on. The best picture available so far on this subject is this wonderful map, of taxa over the skin across the entire body courtesy of the NIH:
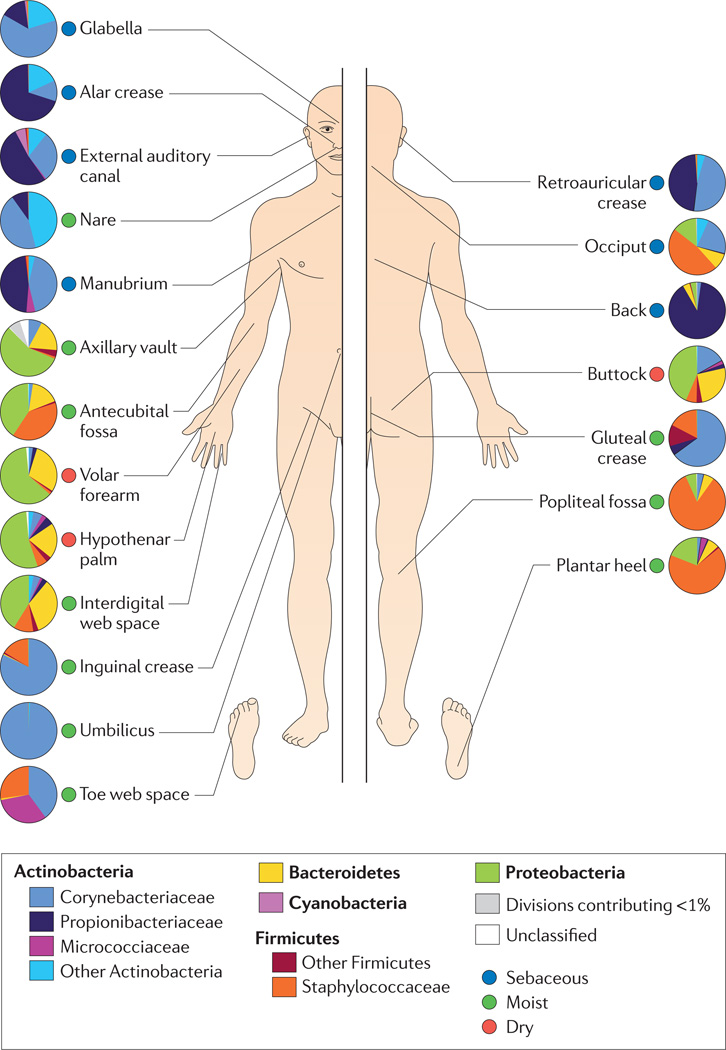
Pause for a moment and look at the differences in composition present in even very close areas. To your microbes there is a world of a difference between your heel and your toes. Microbes operate at a different scale, sensitive to changes so minute you would need a microscope to observe them. Your lower back and upper back. Your neck versus your shoulders. Each one possesses its own distinct ecosystem, each one a unique home for a different microbiome.
Time too is a factor. As your body ages, it transforms, gradually shifting the nutrients and support it provides to its microbiome. Massive microbial drop-off is common as people near retirement age.

Image reproduced from: Front Cell Infect Microbiol. 2012; 2:
- Published online Aug 9, 2012. doi: 10.3389/fcimb.2012.00104 Copyright/License: Copyright (c) 2012 Ottman, Smidt, de Vos and Belzer. Used with permission under the terms of the Creative Commons Attribution License
Original Legend: Human microbiota: onset and shaping through life stages and perturbations. The graph provides a global overview of the relative abundance of key phyla of the human microbiota composition in different stages of life. Measured by either 16S RNA or metagenomic approaches (DNA). Data arriving from: Babies breast- and formula-fed (Schwartz et al., 2012), baby solid food (Koenig et al., 2011), toddler antibiotic treatment (Koenig et al., 2011), toddler healthy or malnourished (Monira et al., 2011), adult, elderly, and centenarian healthy (Biagi et al., 2010), and adult obese (Zhang et al., 2009).
You might want to read the full paper.
The Role of Your Microbiome in Your Body
Reminder: This is a tour of the scientific literature, not medical advice or an alternative to the opinion of your doctor. Please behave responsibly and use it for intellectual appreciation, not self-treatment.
Having a properly functioning microbiome is similar to getting enough sleep – many systems rely on it in complex, hard-to-understand ways. We know you need it from seeing what happens when it’s missing, but the details of why you need it are hard to tease out with current scientific tools. This section should not be taken to contain final answers. It summarizes current research, some of which may turn out to not be reproducible, or to contain perspectives that will not stand the test of time.
Allergies and the Immune System
An estimated 50 million Americans suffer from allergies, which occur when the immune system becomes hypersensitive and reacts against otherwise harmless substances. Heredity and other factors have long been known, but current research suggests that childhood allergy development is related to the child’s microbiota.
Papers
Sjogren (2009) found that children colonized during their first 2 months with Lactobacilli Group I are less likely to develop allergies.
Johansson et al. (2011), in their follow up study, found that inheritable factors for allergies can have an effect on the gut microbiome, but it is not the only factor on the risk of allergy. Early Lactobacilli (L. casei, L. paracasei, L. rhamnosus) colonization also seems to decrease the risk for allergy at five years of age despite allergic heredity.
Stefka et al. (2014) have results that suggest that antibiotic-related microbiome depletion contributed to the 18% increase in food allergies among children in the United States between 1997 and 2007.
Abstract: Environmentally induced alterations in the commensal microbiota have been implicated in the increasing prevalence of food allergy. We show here that sensitization to a food allergen is increased in mice that have been treated with antibiotics or are devoid of a commensal microbiota. By selectively colonizing gnotobiotic mice, we demonstrate that the allergy-protective capacity is conferred by a Clostridia-containing microbiota. Microarray analysis of intestinal epithelial cells from gnotobiotic mice revealed a previously unidentified mechanism by which Clostridia regulate innate lymphoid cell function and intestinal epithelial permeability to protect against allergen sensitization. Our findings will inform the development of novel approaches to prevent or treat food allergy based on modulating the composition of the intestinal microbiota.
Rheumatoid Arthritis
RA is a chronic, disabling, currently incurable and poorly understood autoimmune disease. There is new evidence that suggests that the microbiome plays an important part in this, with both a certain fungus and a certain bacterium identified as triggers for this disease in mouse models. The current view suggests that the presence of a particular microbiome can trigger the disease in genetically predisposed individuals.
Papers
Abstract: (...) Although currently categorized as an autoimmune disorder and regarded as a complex genetic disease, the ultimate cause of rheumatoid arthritis (RA) remains elusive. It seems that interplay between predisposing genetic factors and environmental triggers is required for disease manifestation. New insights from DNA sequence-based analyses of gut microbial communities and a renewed interest in mucosal immunology suggest that the microbiome represents an important environmental factor that can influence autoimmune disease manifestation. This Review summarizes the historical clues that suggest a possible role for the microbiota in the pathogenesis of RA, and will focus on new technologies that might provide scientific evidence to support this hypothesis.
From the conclusions: A fine equilibrium between ‘peace-keeping’ and potentially proinflammatory intestinal bacteria is necessary to keep gut immunity in check and prevent a state of dysbiosis, which might lead to local and distant deleterious consequences in the host. Impressive advances in sequencing technologies, compelling animal data and mounting human evidence suggest that gut microbiota indeed play a part in the pathogenesis of diseases such as autoimmune arthritis ...
Yoshitomi et al 2005 found that the injection of a certain fungal compound could induce RA in normal mice (transient arthritis) and in SKG mice (severe chronic arthritis). Also, treating arthritis-prone mice with an antifungal agent can prevent RA in the mouse model.
Thus, specific microbes, including fungi and viruses, may evoke autoimmune arthritis such as rheumatoid arthritis by stimulating innate immunity in individuals who harbor potentially arthritogenic autoimmune T cells as a result of genetic anomalies or variations.
Abstract: Commensal microbes can have a substantial impact on autoimmune disorders, but the underlying molecular and cellular mechanisms remain largely unexplored. We report that autoimmune arthritis was strongly attenuated in the K/BxN mouse model under germ-free (GF) conditions, accompanied by reductions in serum autoantibody titers, splenic autoantibody-secreting cells, germinal centers, and the splenic T helper 17 (Th17) cell population. Neutralization of interleukin-17 prevented arthritis development in specific-pathogen-free K/BxN mice resulting from a direct effect of this cytokine on B cells to inhibit germinal center formation. The systemic deficiencies of the GF animals reflected a loss of Th17 cells from the small intestinal lamina propria. Introduction of a single gut-residing species, segmented filamentous bacteria, into GF animals reinstated the lamina propria Th17 cell compartment and production of autoantibodies, and arthritis rapidly ensued. Thus, a single commensal microbe, via its ability to promote a specific Th cell subset, can drive an autoimmune disease.
Brain and Nervous System
Alzheimer’s
This area is under active investigation, so the connections aren’t completely clear. But the early evidence suggests that the microbiome might be one of the factors involved in Alzheimer’s. Furthermore, some reviews suggest the possibility of a microbiome-based therapy ( Forsythe et al. 2012 and Collins et al. 2013).
Papers
Depression
Another important link is between microbes and clinical depressive episodes. Microbes seem to influence the regulation of the hypothalamic-pituitary-adrenal (HPA) axis. Messaoudi et al, 2011 did experiments on mice that suggest that a probiotic can reduce depression and psychological distress, among other effects. Another way that depression and stress levels may be related is through the inflammation response
Memory
In 2011, Melanie Gareau et al. found that mice without microbes had impaired memory.
They ran an experiment in which they had:
- mice without microbes and not subject to stress
- mice without microbes and subject to stress
- mice infected with a pathogenic microbe and not subject to stress
- mice infected with a pathogenic microbe and subject to stress
- mice with normal microbes (control group)
The mice without microbes had impaired memory formation, regardless of stress. Mice infected with a pathogen and not subject to stress did not show behavioral anomalies. But, mice infected with a pathogenic microbe subject to stress had memory dysfunctions 10 and 30 days after infection. Daily probiotic treatment of these mice restored their brain function. Another remarkable result is the fact that the mice treated with probiotic not only had their memory function restored, but also had a decreased swelling of cells, compered to mice infected and stressed but not treated with probiotics. (Gareau et al. 2011)
To a certain degree, this is a bit of an academic finding, since most people will have microbes of some kind or another. On the other hand, it is illustrative of the dramatic effect that our symbiosis can have on our daily living. Even more, it illustrates therapeutic avenues that we hope are further explored.
Papers
A pop science summary of Gareau’s work
Gut
CDI/CDAD - Clostridium difficile (C. difficile) infections
C. difficile infections are the poster child of how important microbes are, and of the long term costs of microbial imbalance.
Many healthy individuals have C. difficile in their stomachs. When your digestive ecosystem is healthy, C. difficile has a lot of competition. When the digestive ecosystem is destabilized (for example, through antibiotic treatment), C. difficile can grow to take over your stomach leading to copious diarrhea and, in 10% of cases, death. This happens because C. difficile is resistant to modern antibiotics.
From the CDC page on the subject :
People getting medical care can catch serious infections called healthcare-associated infections (HAIs). While most types of HAIs are declining, one – caused by the germ C. difficile – remains at historically high levels. C. difficile causes diarrhea and is linked to 14,000 American deaths each year. Those most at risk are people, especially older adults, who take antibiotics and also get medical care. (...)
The two best antibiotics for the treatment of C. difficile are vancomycin and fidaxomicin. Treatment has a success rate of around 75%, fidaxomicin is designed specifically for C. difficile but it is quite expensive and not typically the first line treatment, and is not fully clear that is the best option (Bartsch et al., 2013).
Intriguingly, there is also a microbial treatment: recolonizing weakened gut flora with the gut flora of a healthy donor. It’s called FMT (fecal microbiota transplant). Although it has an unpleasant name, the treatment is economical, and is effective on about 90% of the time (Bakken et al, 2012).
From Rohlke & Stollman (2012):
Abstract: Clostridium difficile infection rates are climbing in frequency and severity, and the spectrum of susceptible patients is expanding beyond the traditional scope of hospitalized patients receiving antibiotics. Fecal microbiota transplantation is becoming increasingly accepted as an effective and safe intervention in patients with recurrent disease, likely due to the restoration of a disrupted microbiome. Cure rates of > 90% are being consistently reported from multiple centers. Transplantation can be provided through a variety of methodologies, either to the lower proximal, lower distal, or upper gastrointestinal tract. This review summarizes reported results, factors in donor selection, appropriate patient criteria, and the various preparations and mechanisms of fecal microbiota transplant delivery available to clinicians and patients.
In summary: A healthy microbiota protects you by outcompeting C. difficile.
Links:
Medscape article on cost efficacy of FMT
Crohn’s disease / IBS
The current view is that it seems likely that the microbiome might be a marker for Chrohn’s disease Erickson et al, 2012, and furthermore it is also suspected to be a causal factor in at least some of the cases. The effect of the microbiota on Chrohn’s is potentially mediated by the immune system. However, research on the relationship between Chrohn’s and the microbiome is still preliminary.
Some of the findings we have so far are:
Conclusions: The metagenomic approach allowed us to detect a reduced complexity of the bacterial phylum Firmicutes as a signature of the faecal microbiota in patients with CD. It also indicated the presence of new bacterial species.
Discussion: This study describes, for the first time, the fecal microbiome profile of a large number of children hospitalized with severe UC using a comprehensive human fecal microbial 16S rRNA microarray. We found substantial reductions in the richness, evenness, and biodiversity of the gut microbiome in these children, supporting a correlation between microbial diversity and bowel health. Although data on disease severity were available for each individual, the number of patients was too small to directly correlate severity with microbial diversity. However, the fact that patients refractory to corticosteroids had further reduction in microbial richness, relative to those who responded to therapy, suggests a “dose response” effect, where more severe illness associates with further reduced richness. Our findings cannot determine whether poor diversity is a cause or result of severe colitis; however, this feature could be used as an important marker of severity, possibly even predicting response to corticosteroids.
Colon Cancer
Current understanding suggests your microbiome might be a marker of the presence of colon cancer (Zackular et al, 2013,Baxter et al., 2014). In particular, Fusobacterium has been implicated as being present in colon carcinomas, while at the same time Bacteriodetes and Firmicutes are depleted (Kostic et al., 2012).
Whether the microbiome is a causal agent or one of the factors in colon cancer is something on which there is no clear consensus yet. A review paper by Tjalsma et al (2012) proposes a model for the effect of the microbiome in the development of colorectal cancer.
Kwashiorkor
Kwashiorkor is a form of acute malnutrition, and it seems to be caused by environmental factors and a lack of protein in the diet. Recently, the team of Prof. Gordon at WUSTL established that the microbiome is one of the causal factors in this disease.
This is one of the few cases in which we have advanced enough to have established a “bad microbiome” to be one of the causal factors of a disease. Granted, it is not the only factor, but it is one of them rather than just a marker of the disease.
General links
Papers
Smith et al., 2014, Gut microbiome of Malawian twin pairs discordant for kwashiorkor.
Abstract: Kwashiorkor, an enigmatic form of severe acute malnutrition, is the consequence of inadequate nutrient intake plus additional environmental insults. To investigate the role of the gut microbiome, we studied 317 Malawian twin pairs during the first 3 years of life. During this time, half of the twin pairs remained well nourished, whereas 43% became discordant, and 7% manifested concordance for acute malnutrition. Both children in twin pairs discordant for kwashiorkor were treated with a peanut-based, ready-to-use therapeutic food (RUTF). Time-series metagenomic studies revealed that RUTF produced a transient maturation of metabolic functions in kwashiorkor gut microbiomes that regressed when administration of RUTF was stopped. Previously frozen fecal communities from several discordant pairs were each transplanted into gnotobiotic mice. The combination of Malawian diet and kwashiorkor microbiome produced marked weight loss in recipient mice, accompanied by perturbations in amino acid, carbohydrate, and intermediary metabolism that were only transiently ameliorated with RUTF. These findings implicate the gut microbiome as a causal factor in Kwashiorkor.
Obesity and metabolic syndrome
Obesity / metabolic syndrome is a cluster of multiple conditions coming together to increase the risk of associated complications (such as diabetes, heart disease and stroke), and it’s a complicated condition.
So far, the scientific jury is still out on what exactly is happening with the microbiome in obesity / metabolic syndrome. Yet we know that microbes are definitively part of the picture. One of the first results in the field was the association of a lean microbiome with lean mice and an obese microbiome with obese mice.
At first, it seemed like they were clearly different, and transplanting them would make the obese mice become lean (Turnbaugh, 2006) and, if translatable to humans, solve the problem. They also found a relative abundance of Firmicutes and a relative scarcity of Bacteroidetes in obese mice when compared to lean mice (Ley et al, 2005, Ley et al., 2006) and suggested that manipulating the microbial community would allow regulation of the energy balance in obese humans.
Later, Schwiertz et al. (2010) found that there were plenty of Bacteriodetes in overweight people. One thing they agreed with in Gordon’s previous work was the role of short chain fatty acids in the microbiota of obese people (they tend to have more of the short chain fatty acid metabolism).
A follow up study by Jumpertz et al. (2011), looked at the diet of both obese and lean people. This study employed two groups, one made of lean people eating 2,400 calories per day and another of obese people eating 3,500 calories per day. When people ate a weight-maintining diet (2,400 calories per day), no differences in the bacterial abundance could be found between the lean and obese group. For the group on the 2,400 calories diet, their Firmicutes became more abundant to increase the energy intake. The group of obese people did not exhibit any clear-cut pattern. This could have been due to the small group size, underlying genetic factors, or something else.
But it seems that microbes definitively matter, although the details are still under study.
Another interesting study is the one by Vijay-Kumar et al. (2010). They used a special type of mouse that lacks the ability to recognize TLR-5, and this seems to result in something that resembles human metabolic syndrome (Vijay-Kumar et al., 2010).
Abstract: Metabolic syndrome is a group of obesity-related metabolic abnormalities that increase an individual's risk of developing type 2 diabetes and cardiovascular disease. Here, we show that mice genetically deficient in Toll-like receptor 5 (TLR5), a component of the innate immune system that is expressed in the gut mucosa and that helps defend against infection, exhibit hyperphagia and develop hallmark features of metabolic syndrome, including hyperlipidemia, hypertension, insulin resistance, and increased adiposity. These metabolic changes correlated with changes in the composition of the gut microbiota, and transfer of the gut microbiota from TLR5-deficient mice to wild-type germ-free mice conferred many features of metabolic syndrome to the recipients. Food restriction prevented obesity, but not insulin resistance, in the TLR5-deficient mice. These results support the emerging view that the gut microbiota contributes to metabolic disease and suggest that malfunction of the innate immune system may promote the development of metabolic syndrome.
Links
Papers
-
Jumpertz et al., 2011, Energy-balance studies reveal associations between gut microbes, caloric load and nutrient absorption in humans.
-
Ley et al., 2006, Microbial ecology: human gut microbes associated with obesity.
-
Schwiertz et al.,2010, Microbiota and SCFA in lean and overwight healthy subjects.
-
Turnbaugh et al., 2006, An obesity-associated gut microbiome with increased capacity for energy harvest.
-
Turnbaugh et al., 2008, A core gut microbiome in obese and lean twins.
Sign post: For an overview of Equilibrium’s design, please see the product page and scroll down. We also welcome questions at questions@generalbiotics.com.